In-Silico Investigation of Plant-Derived Natural Allosteric Compounds Towards Enhanced Drug-Protein Interaction of MOA Protein Complex in Depression Based on Molecular Docking and Molecular Dynamic Simulation Approaches
DOI:
https://doi.org/10.5530/ctbp.2022.4.86Keywords:
Allosteric compound, Molecular docking, Homaline, Major Depressive Disorder,, Molecular dynamic simulationsAbstract
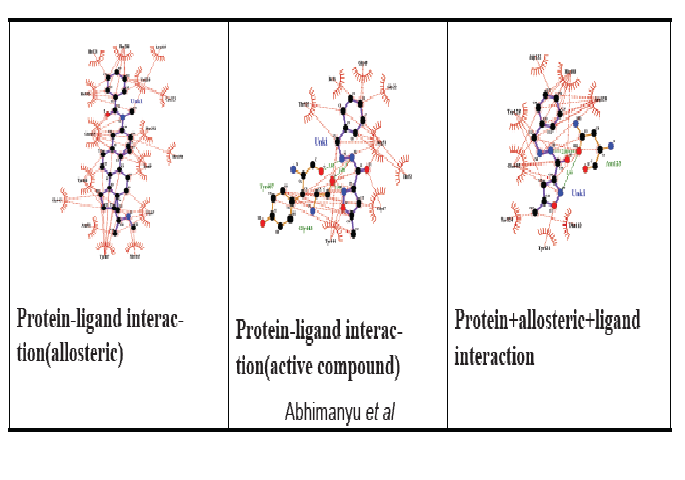
Allostery is an effective method of controlling the function of biological macromolecules and currently gaining more attention in the realm of drug development due to the unique characteristics of allosteric modulators, such as good selectivity and low toxicity. These qualities are critical for both the creation of the allosteric concept and the evaluation of allosteric interactions. Primarily, allosteric modulators are responsible for boosting efficacy and reducing the drug’s catastrophic effects. Since the chemical compounds have always proven to cause side effects in the body. Hence, the discovery of an alternative natural allosteric compound instead of chemical allosteric compounds for enhancement of drug efficacy and regulating drug dosage is the major challenge in modern drug discovery, especially for diseases i.e., cancer, Parkinson’s, mental disorder, etc. where the long-term treatments are recommended. In this research paper in-silico based comparative interaction/molecular docking and dynamics study of FDA-approved Antidepressant drugs i.e., isocarboxazid Phenelzine, Selegiline, Tranylcypromine, etc with potential drug target protein MAO enzymes for major depressive disorder have been conducted. Various drug compounds complexed with protein have been analyzed by performing molecular docking. The site-directed docking and interaction energy for the Isocarboxazid drug complexed with protein target was performed with a docking score -8.6 kcal/mol using Autodockvina. Read more . . .