Identification of Potential Inhibitors Against Mycobacteriaum tuberculosis Protein Targets: An in silico Approach
DOI:
https://doi.org/10.5530/ctbp.2022.2s.33Keywords:
Drug targets, Mycobacterium tuber- culosis, Lipinski’s rule, Molecular docking, Ser- ine/threonine-protein kinase PknDAbstract
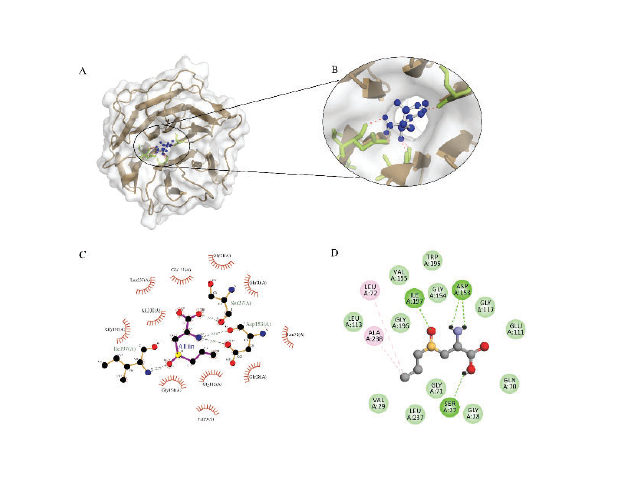
Mycobacterium tuberculosis (Mtb) is gradually gaining resistance to anti-tuberculosis drugs, which have imposed novel drug targets on Mtb for further discovery. Seven proteins involved in the cellular processes of Mtb with their respective PDB IDs, namely rpoC (5UH5), PstS3 (4LVQ), ICDH-1 (4HCX), cpn10 (1P3H),PstS1 (1PC3), PknD (1RWI) and devR/dosR (3C3W), were obtained from the RCSB Pro- tein Data Bank (PDB) and thoroughly analyzed. Literature curated 80 compounds were virtual- ly screened using various bioinformatics tools such as Molinspiration, Osiris Property Explor- er, ProTox-II. Molecular docking interactions of Mtb-Serine/threonine-protein kinase PknD with Alliin ligand shows better binding energy (-6.07 kcal/mol) and the specific key residues such as SER27, GLY28, VAL29, GLN70, GLY71, LEU72, GLU111, GLY112, ASP153, GLY154,GLY196, ILE197, and ALA238 play a vital role in ligand interaction using hydrogen bonds, van der Waals and hydrophobic interaction. Therefore, the finding shows alliin is the ideal phyto-compound that could be used for the op- timized synonymous compound design for safe and efficient tuberculosis treatment specific to the PknD protein target.