A Comparative In Silico Analysis on Frequency and Distribution of Microsatellites in Whole Genome Sequences of Three Pathogenic Candida species and Development of SSR markers for Diversity Analysis
DOI:
https://doi.org/10.5530/ctbp.2022.2.18Keywords:
Candida, Microsatellites, polymorphic markers, Genetic diversityAbstract
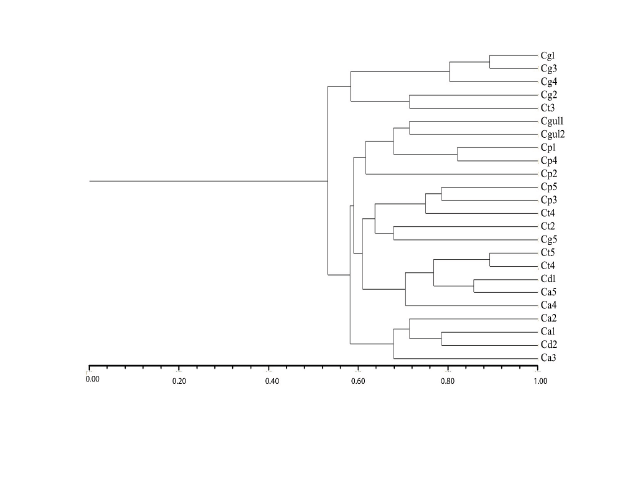
The species of the genus Candida are well known for their ability to cause fungal colonization and infection in humans, which is commonly known as candidiasis. Among them, Candida albicans, Candida glabrata, Candida parapsilosis, and Candida tropicalis are reported to be frequently occurring pathogenic species, which are responsible for superficial and systemic infection. In the present study, we surveyed the microsatellites in the available whole-genome sequences of three pathogenic species viz. C.albicans, C.glabrata and C.tropicalis. The relative abundance and density was higher in C.albicans when compared to C.glabrata and C.tropicalis. Thirty microsatellite primers were designed, ten from each species, for genetic characterization of Candida isolates belonging to six Candida species. Of the 30 markers, only nineteen showed amplifications. A total of 32 alleles were amplified by 19 primers with an average of two alleles per marker. Ten markers showed 100% polymorphism. The markers were found to be more polymorphic (75%) in C.albicans as compared to C.glabrata and C.tropicalis, however, polymorphic information content was the maximum (0.75) in CgSSR 4. Twelve polymorphic markers obtained in this study clearly demonstrate the utility of newly developed markers in establishing genetic relationships among different isolates of Candida.