An Integrative Bioinformatics Analysis for Identifying Hub Genes in Human Immunodeficiency Virus
DOI:
https://doi.org/10.5530/ctbp.2021.6.13Keywords:
HIV, Microarray analysis, PPI interactions, Functional annotation, Pathway analysis, Topological analysisAbstract
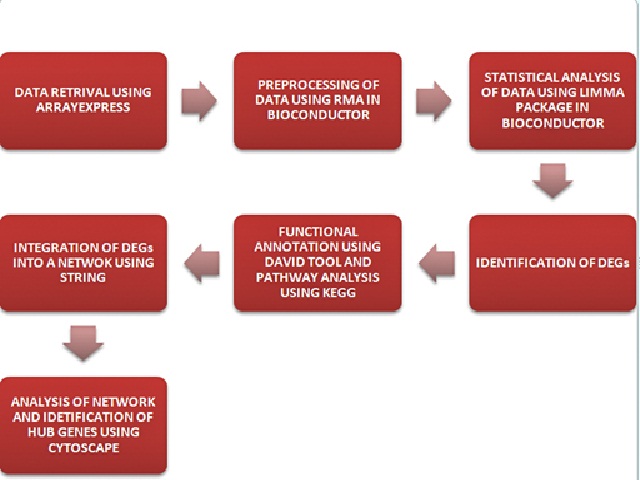
The inability of modern medicine to find a cure for human immunodeficiency virus (HIV) since its discovery as the causative agent of acquired immunodeficiency syndrome (AIDS), has made HIV as one of the most dreadful pathogens of this century. The present study presents the putative drug targets and biomarkers of HIV using bioinformatic tools. Microarray data analysis of datasets that involved HIV cases vs controls was done in order to scrutinize the differentially expressed genes (DEGs). From this data a total of 57 common up-regulated DEGs and 108 common down-regulated genes were obtained for HIV. The functional annotation and pathway analysis of the DEGs was done in order to get the biological processes and pathwaysin which the DEGs were enriched. From the analysis of the network the significant genes that were obtained for HIV datasets were ‘Protein Kinase C Alpha (PRKCA)’, ‘Signal Transducer and Activator of Transcription 5B (STAT5B)’, ‘Krupple Like Factor-6 (KLF6)’, ‘Granzyme K (GZMK)’, ‘T-cell Immunoreceptor With Ig and ITIM Domains (TIGIT)’, ‘Ectonucleoside Triphosphate Diphosphohydrolase 1 (ENTPD1)’, ‘Regulator of G protein signaling 1 (RGS1)’, ‘CD48 Molecules (CD48)’, ‘Cytotoxicity and Regulatory T-cell Molecule (CRTAM)’, and ‘Neutrophil Cytosolic factor 4 (NCF4)’. The topological analysis was done for these genes for better understanding of their interaction.
The present study also suggests that the significant DEGs and Biological processes and pathways that accompany them might have the potential to be exploited as possible drug targets and biomarkers in the diagnosis, prognosis as well as treatment of HIV and its comorbidities and warrants for further experimental validation.