Molecular Docking and MD Simulation Analysis of L-asparaginase of Streptomyces iranensis, Streptomyces himalayensis and Streptomyces griseus
DOI:
https://doi.org/10.5530/ctbp.2021.4.40Keywords:
AutoDock Vina,, Cabs Flex, CB-Dock, I-TASSER, StreptomycesAbstract
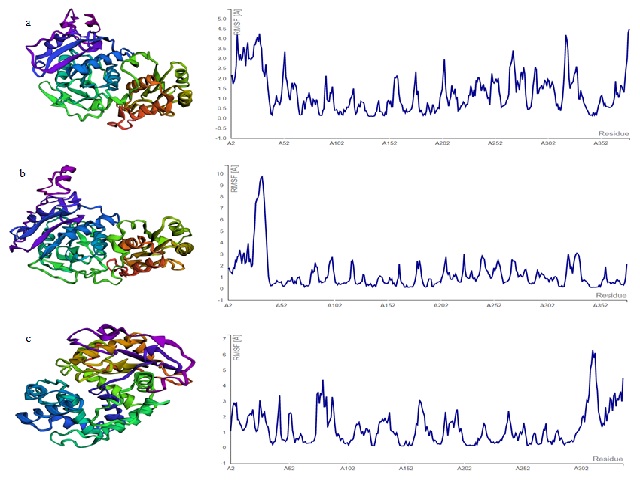
The present study focuses on structural, molecular docking and simulation analysis of L-asparaginase protein belonging to Streptomyces iranensis (S. iranensis) (WP_044568095), Streptomyces himalayensis (S. himalayensis) (WP_181864635) and Streptomyces griseus (S. griseus) (WP_037680047). The three protein sequences retrieved from NCBI are also assessed for characteristic, secondary structure and localization studies. The analysis with BLAST tool predicted WP_044568095 to be 40.18% and 39.16% identical with D. chrysanthemi (P06608) and E. coli L-asparaginase II (P00805). Similarly WP_181864635 is identified to be 38.39% and 37.58% identical with P06608, P00805 respectively while no resemblance is observed with WP_037680047.1. The three Streptomyces proteins on subsequent studies by I-TASSER and GalaxyRefine are found to have 85.9%, 84.2% and 79.2% residues in the favoured zone for WP_044568095, WP_181864635 and WP_037680047 on validation by PROCHECK. Further, acceptable findings are obtained from studies by ERRAT and ProSA. Molecular docking performed by CB-Dock and PyRx resulted in similar predictions about the active site amino acids forming hydrogen bond with L-asparagine. WP_044568095 docked with Asn resulted in lowest energy of - 4.6 kcal/mol while - 4.4 kcal/mol is obtained for WP_181864635, WP_037680047 evaluated by AutoDock Vina of PyRx software. The docked structures of WP_044568095 and WP_181864635 under controlled conditions by CABS-Flex 2.0 are found stable in comparison with WP_037680047. As L-asparaginase possess high therapeutic application in various malignancies treatment, the evaluated proteins of S. iranensis, S. himalayensis and S. griseus are worth analysing further for in vitro enzyme activity and are considered to have promising therapeutic importance.