A Preliminary Bioinformatics Data Analysis of Single Nucleotide Polymorphisms of the PON1 Genes in Chronic Kidney Disease: In Silico Analysis
DOI:
https://doi.org/10.5530/ctbp.2025.2.14Keywords:
In silico, Single nucleotide polymorphisms, Serum paraoxonase (PON1), chronic kidney disease (CKD)Abstract
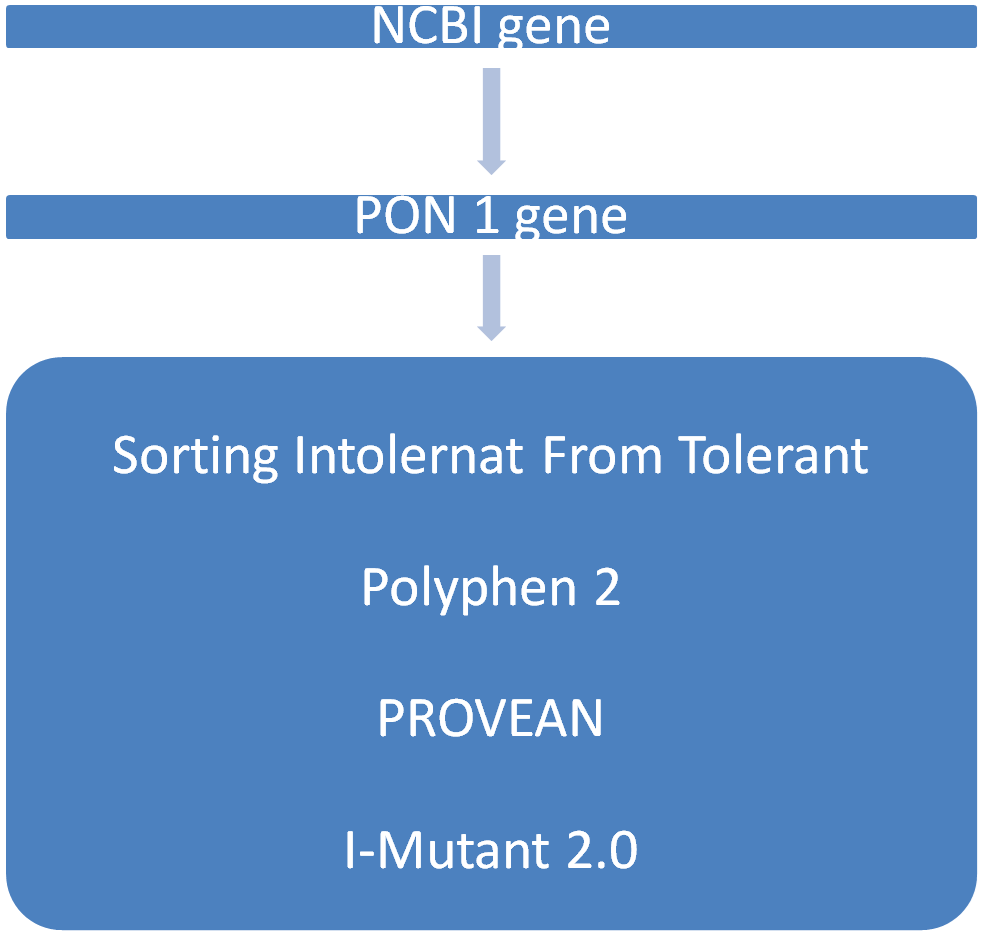
Serum paraoxonase (PON1), a glycoprotein synthesized in the liver, protects against oxidative stress and lipid peroxidation, potentially reducing the risk of chronic kidney disease (CKD). A study using bioinformatics methods, such as PROVEAN (Protein Variation Effect Analyzer), SIFT (Sorting Intolerant from Tolerant), Polyphen 2, and I-Mutant 2.0 analyzed non-synonymous single nucleotide polymorphisms (SNPs) of the PON1 gene. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis and Gene Ontology (GO) enrichment were used to identify biological processes and pathways. SIFT analysis of the PON1 gene’s SNPs showed that 55 and 33 were tolerable, and 22 were harmful alterations. According to PROVEAN analysis, 22 mutations were neutral, and 33 were harmful. Polyphen 2 revealed that 26 were damaging and 32 were benign. Thirty-four SNPs on I-Mutant analysis showed decreased thermodynamic stability, while twenty-one showed enhanced stability. The study found that the structure and function of the PON1 gene are impacted by mutations, with decreased stability predicted. These mutations may affect CKD’s pathobiology and risk for cardiovascular disease. A wet lab investigation on PON1 pathways could help link CKD pathophysiology and progression.