Computational Evaluation of Curcumenol as a Potential Inhibitor against Calcineurin Protein
DOI:
https://doi.org/10.5530/ctbp.2024.4s.6Keywords:
Natural inhibitor, Molecular Docking, Molecular dynamics simulation, Inhibitory concentration 50Abstract
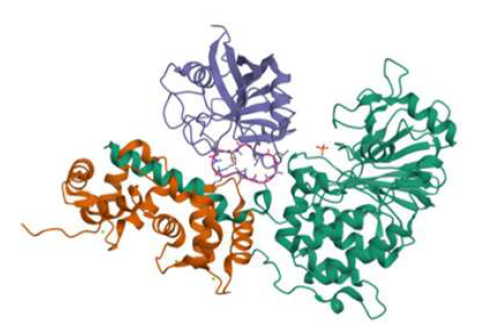
Calcineurin protein is a multifunctional serine-threonine protein phosphatase vital to several cellular functions, such as immune response control, signal transduction, and cardiac function. Its relevance in preserving cellular homeostasis is highlighted by its involvement in T-cell activation, calcium signalling pathways, etc. The dysregulation of the calcineurin protein has been reported to be linked with neurodegenerative diseases, and hence calcineurin inhibitors may be beneficial as a treatment option. Existing FDA-approved calcineurin inhibitors are used under stringent guidelines and require careful monitoring because of possible side effects and toxicity. Consequently, as a therapeutic option, the discovery and development of novel calcineurin inhibitors is imperative. For this reason, the current study aims to characterize the calcineurin inhibitory action of curcumenol, a naturally occurring chemical by employing diverse in-silico techniques. Curcumenol was checked for its ADME-T and molecular interactional stability and was predicted to have impressive inhibitory potential with a docking score of -9.1kcal/mol and a mean RMSD of the backbone atoms of the Calcineurin-curcumenol complex as 4.55Å when simulated in a molecular dynamics system for 100ns. Theinsilicofindings were further justified using in vitro Calcineurin Phosphatase activity assayto quantitively analyse the inhibitory efficacy of curcumenol, defined in terms of IC50 value,which was found to be 0.113 μM. The overall study suggested that curcumenol holds the characteristic molecular and interactional properties and hence can be developed as a prospectiveand safe calcineurin inhibitor which can be ultimately employed as a promising therapeutic candidate against neurodegenerative diseases.