Molecular Dynamics Study of Quinazoline Compounds Complexed with Filamenting Temperature-Sensitive Z Protein and Gyrase Subunit B as Potential Antibacterials
DOI:
https://doi.org/10.5530/ctbp.2024.3s.14Keywords:
Molecular dynamics, Quinazoline derivatives, Filamenting Temperature Sensitive Z Protein, DNA gyrase subunit B, AMBERAbstract
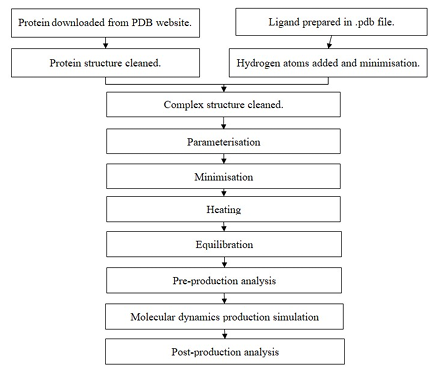
As the years go by, bacteria such as Staphylococcus aureus and Mycobacterium tuberculosis have developed resistance towards the current antibiotics which leads to ineffectiveness of antibacterial agents to kill or inhibit the bacteria. Thus, in order to overcome this issue, quinazoline derivatives have been proposed as the potential new antibacterial agents due to their antibacterial properties. Molecular dynamics, a RMSD (1.2Å for Q44 and 2.6Å for 3U2D), and a higher negative value of free binding energy (-23.21 kcal/mol) with favourable hydrophobic interaction. However, Q44 does not form a significant hydrogen bond as the occupancy is nearly zero. Q100 and Q44 have the most potential quinazoline derivative to act on the FtsZand GyrBrespectivelyto continue to the next step in drug design as a new antibacterial drug candidate. computational technique, has been conducted in this study to determine the potential of quinazoline compounds as a novel antibacterial agent for Staphylococcus aureus’s DNA gyrase subunit B (GyrB) and Mycobacterium tuberculosis’s Filamenting temperature-sensitive Z (FtsZ) protein. Molecular dynamics simulation of the top 2 docked ligands of quinazoline for each FtsZ and GyrB were conducted by using Amber22 molecular dynamics simulation software. The analyses were conducted with cpptrajto evaluate the stability and binding interaction of the compounds with the target receptors.The dynamic studies of Q100 complexed with FtsZ show it is the most stable, with lower RMSD values (1.4Å for Q100 while FtzZ is 2.3Å) and lesser overall variation in RMSF. Although Q100 does not form a significant hydrogen bond with FtsZ, it has a higher negative free energy binding value (-25.48 kcal/mol) compared to Q56 with favourable hydrophobic and electrostatic interaction. While Q44 also shows the complex with GyrB is slightly more stable, with lower RMSF in residue 1 (3.0Å), stable