Suppression of Hotair Gene in Small-Cell Lung Cancer Effect The Differentially Expressed Genes and Inhibit Progression of the Disease
DOI:
https://doi.org/10.5530/ctbp.2024.3s.10Keywords:
Small-cell lung cancer, HOTAIR gene, differentially expressed gene, pathway analysis, cellular mechanismAbstract
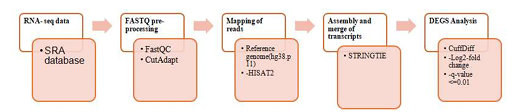
HOTAIR gene is a long non-coding RNA (lncRNA) that regulates the differentially expressed genes causing intermediate malignant respiratory disorders like small-cell lung carcinoma (SCLC). Thus, HOTAIR gene can be a predictor for chemotherapy response and therapeutic target against chemoresistance in SCLC. In this study, SCLC and HOTAIR suppressed genes were compared with normal lung tissue samples (control) to identify the differentially expressed genes (DEGs). The number of significant DEGs in both SCLC and HOTAIR suppressed vs control analyses were 4906 and 255 respectively with a cutoff value of (q≤ 0.05). Among the DEGs, 48 genes were expressed in both SCLC and HOTAIR suppressed conditions with log2 fold change <2. Eight genes (n=8) (ASAH1, CENPE, FABP5, ACOXL, CTSB, RAB11FIP1, VEGFA, JUN) were significantly expressed (p<0.05) in both conditions. CENPE, VEGFA and JUN were lowly expressed in SCLC but highly expressed in HOTAIR suppressed condition. Enrichment analysis on the selected genes using Kyoto Encyclopedia of Genes and Genomes (KEGG) tool shows the cell adhesion and cell cycle pathways were significantly activated in both SCLC and HOTAIR suppressed conditions. About 58 reactome pathways were identified and the post translational protein modification pathway was activated in SCLC while the innate immune system pathway in HOTAIR suppressed gene. These potential HOTAIR suppressed DEGs are CENPE, VEGFA and JUN and they could serve as potential biomarkers for cancer drug development. However, further studies are required to develop the protein interaction model of the enriched genes to analyze the correlation and insight of the therapeutic intervention.