An In-Silico Approach Towards Drug Discovery Against Bacterial DNA Gyrase Using Nalidixic Acid
DOI:
https://doi.org/10.5530/ctbp.2024.4.45Keywords:
Bacterial DNA Gyrase, Drug discovery, oral cancer., Molecular docking interactionAbstract
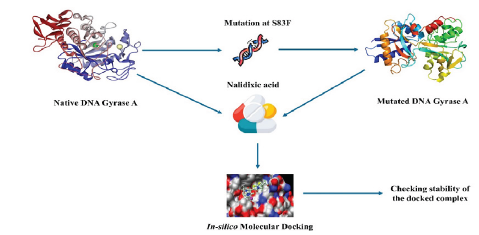
Escherichia coli is a gram- negative bacteria and is common for bacterial disease that affects the intestinal tract. DNA gyrase A was an enzyme which is responsible for DNA breakage and reunion are mostly released by Escherichia coli bacteria in human gastrointestinal tract, causing a situation called gastroenteritis. Normally in native DNA gyrase A protein, the 83th position is occupied by serine residue which is mutated to phenylalanine making the enzyme as antibiotic resistant against the known antibacterial drugs. Nalidixic acid is mainly used in the treatment for urinary tract infections caused by gram-negative bacteria. Here, according to our present study, we compared binding affinities of native protein structure and mutated protein structure of E. coli DNA gyrase A against the Nalidixic acid. ADMET properties, bioactive scores and other parameters of the ligand structure were calculated using various tools including SwissADME, pkCSM software as well as PreADMET web tool. Lastly molecular docking study was carried out using AutoDock Vina software and the results were evaluated on the basis of iMod server. After comparing the docking scores, it was observed that the mutated protein (-5.5 kCal/mol.) shows more binding affinity towards nalidixic acid than the native protein (-5.3 kCal/mol.) Although, more in-vitro and in-vivo studies need to be performed to get a satisfactory conclusion.