Genetic Impact of Tobacco Smoke on Blood and Airway Epithelium: A Transcriptional Profiling Study
DOI:
https://doi.org/10.5530/ctbp.2024.1.13Keywords:
Smoking, Gene Expression, Protein-Protein network, gene erichmentAbstract
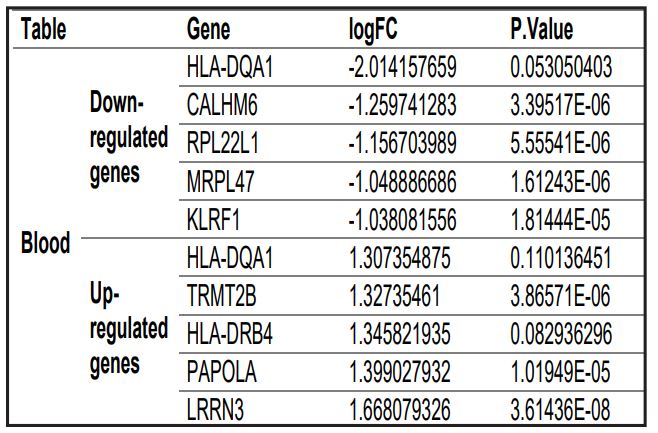
Tobacco smoking poses significant health risks due to its carcinogenic and toxic components. While the broad effects of smoking are well-documented, the specific genomic consequences on different body tissues remain less understood. This study seeks to address this knowledge gap by systematically analyzing the genetic alterations in blood and airway epithelium tissues of smokers, offering a clearer understanding of the molecular pathways impacted by tobacco exposure. In this study, we employed microarray-based gene expression analysis to investigate the genomic changes in smokers. Employing gene expression datasets (E-MTAB-5279 and E-GEOD-10006) from smokers and non-smokers, we conducted a detailed analysis to identify differentially expressed genes (DEGs) in blood and epithelial tissues. Our methodology included robust multi-array average processing, Limma package analysis for DEGs, and pathway enrichment analysis using the KEGG database and Gene Ontology (GO) tools. The analysis revealed 584 DEGs in the blood dataset, with 411 downregulated and 173 upregulated genes, highlighting pathways related to thermogenesis, Parkinson’s disease, Alzheimer’s disease, and Huntington’s disease. Key genes such as ATP5C1, ATP5J, COX6C, COX7B, COX7C, NDUFA4, NDUFA5, NDUFB3, NDUFS4, UQCRB, and UQCRQ were notably enriched. In the airway epithelium dataset, 147 DEGs were identified, including 101 downregulated and 46 upregulated genes, with a significant enrichment in the ribosome pathway, particularly in genes like RPL23. In the airway epithelium dataset, we identified 147 differentially expressed genes (DEGs), consisting of 101 downregulated and 46 upregulated genes. Notably, there was a pronounced enrichment in the ribosomal pathway, exemplified by genes such as RPL23. This finding highlights a critical cellular response to tobacco smoke, emphasizing alterations in protein synthesis mechanisms within the epithelial tissues. In conclusion, our findings provide a comprehensive view of the genomic changes induced by tobacco smoke in different tissues, enhancing our understanding of smoking-related pathologies and potentially guiding future therapeutic strategies. This research not only fills a crucial gap in the understanding of tobacco’s genomic impact but also lays the groundwork for improved public health interventions.