Extraction of Extracellular Membrane Vesicles (EMV) from Streptococcus pneumoniae using Ultracentrifugation, Ultrafiltration and Iodixanol Gradient Fractionation
DOI:
https://doi.org/10.5530/ctbp.2023.4s.97Keywords:
Streptococcus pneumoniae, extracellular membrane vesicle, ultracentrifugationAbstract
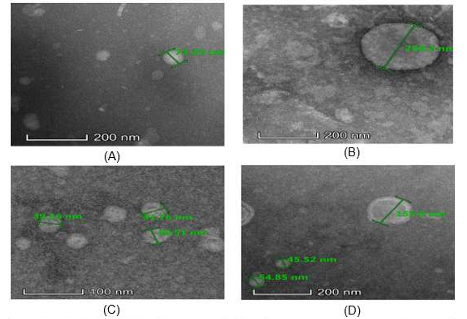
Extracellular membrane vesicles (EMVs) are membranous structures that are excreted by gram-positive bacteria. These vesicles are involved in a multitude of biological functions, essential for adaptability to the environment, cellular component exchange, antigen and virulence factor distribution, and infection transmission. Recently, bacterial EMVs have gained attention due to their potential as highly effective vaccine targets. However, extraction of EMVs from bacterial cells has been difficult, especially among gram-positive organisms. This study aimed to optimize a method to extract EMVs from Streptococcus pneumoniae which can be used as a potential vaccine candidate. The EMVs of S. pneumoniae was extracted from its common serotypes (6A, 14, 19A, 19F, and 23F) using ultracentrifugation, ultrafiltration, and iodixanol gradient fractionation. The extracted EMVs were validated by viewing their morphology using a transmission electron microscope (TEM). The six S. pneumoniae serotypes used were found to release extracellular vesicles, albeit in different numbers and sizes (22 nm - 250 nm). They are believed to contain various biologically active proteins required for bacterial nutrient acquisition, biofilm formation, and pathogenesis. The success of extracting EMVs from S. pneumoniae using a modified method has paved a path to study better drug targets for S. pneumoniae since bacterial EMVs are non-viable components of the bacteria that act as an antigen, hence able to induce host immune response. This suggests EMVs as potential vaccine candidates for this bacterium.